CAP recommends using neoplastic cellularity as the number of neoplastic cells or nuclei compared to the overall or toal number of cells or nuclei.

Massive Parallel Sequencing (Next Generation Sequencing)
References databases:
1. Atlas of Genetics and Cytogenetics in Oncology and Haematology
2. Catalogue of Somatic Mutations in Cancer COSMIC, databases in the Wellcome Sanger Institute.
3. Clinical Interpretation of Variants in Caner CIViC, identifying evidence levels and clinical trials.
4. Franklin, Variant interpretations.
5. Genome Nexus, OncoKB Therapeutic Levels.
6. Mutalyzer, check HGVS nomenclature, position conversion, SNP converter,...
7. NCBI ClinVar and dBSNP, containing only well documented reportable variants.
8. Varsome, assist in following ACMG and AMP classification.
Massive Parallel Squencing (Next Generation Sequencing) is ran on the Illumina iSeq 100, MiniSeq and NextSeq 500, using "sequencing by synthesis" chemistry, upon different targeted sequencing panels.
Genomic DNA QC : Quantified by Qubit and the integrity control by conventional PCR
Library QC : Check by automatic electrophoresis (Bioanalyser)
Sequencing QC : Coverage > 500x, Q30 > 80%
Bioinformatics :
Illumina Local Run Manager on a desktop PC
DNA Amplicon (BaseSpace Workflow) 2.1.0.19 [DNA Amplicon Workflow (3.24.1.8+
master), BWA-MEM Whole-Genome(Aligner)0.7.9a-isis-1.0.2, Pisces
Variant Caller 5.2.9.23, Illumina Annotation Engine 2.0.11-0-
g7fb24a09, Bam Metrics v0.0.22, and SAMtools 0.1.19-isis-1.0.3,
Annotation Dataset 91.26.44]
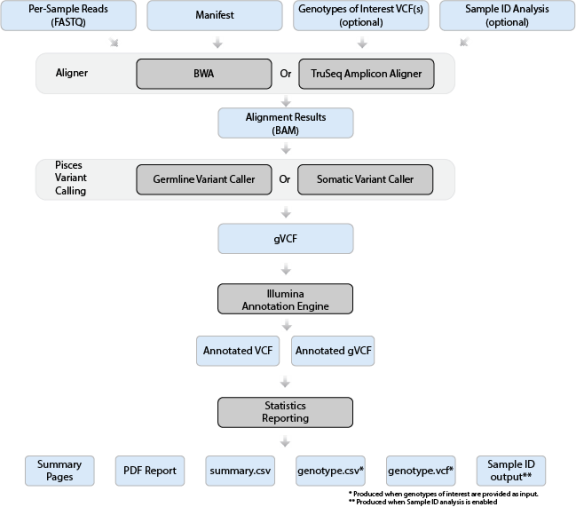
Curation :
Human GRCh37/hg19, ClinVar, Genome Nexus, Franklin, VarSome
Limitations : The workflow could detect SNV-indel type mutations with a variant allelic frequency greater or equal to 5%. However, variants with VAF<10% might fall short of Sanger sequencing confirmation. Due to the nature of FFPE specimen and limitations of the amplicon-based library preparation, genomic changes leading to false negativity, including, but not limited to, large deletion, large duplication and gross chromosomal abnormalities, cannot be completely excluded.
The Ion Torrent Genexus System automates sample and library preparation, sequencing, analysis, and reporting. The Genexus Purification System and Genexus Integrated Sequencer with Genexus Software work together seamlessly, tracking sample information and results automatically through the process. This automated system reduces manual steps and facilitates quality data and quick reports.
Genomic DNA/RNA QC : Quantified by Qubit and the integrity control by conventional PCR
Library QC : Integrated in the automated system
DNA Sequencing QC : >=Q20 bases, Uniformity
RNA Sequencing QC : >=Q20 bases, Mapped
Bioinformatics : Integrated
The factory bioinformatics reports a standardized table of known variants. An in-house bioinformatic pipeline (GATK-Mutect2) is installed for screening uncalled variants for the 12-gene lung cancer panel. Sequencing data of selected cases is also reviewed by Integrated Genome Viewer (IGV).
Curation :
Human GRCh37/hg19, ClinVar, Genome Nexus, Franklin, VarSome
Limitations : The workflow could detect SNV-indel type mutations with a variant allelic frequency greater or equal to 5%. However, variants with VAF less than 10% might fall short of Sanger sequencing confirmation. Due to the nature of FFPE specimen and limitations of the amplicon-based library preparation, false negative results cannot be completely excluded. These latter missed genomic changes might include large deletion, large duplication, gross chromosomal abnormalities, intronic variants, etc.
Somatic and germline testing for BRCA1 and BRCA2 genes will be offered for patients with high grade serous carcinoma for determing elligibility to targeted therapy.
Germline testing for BRCA1 and BRCA2 genes will also be offered for all patients with breast cancers, to assess genetic risk.
Somatic mutation testing is done on the FFPE block or cytology cell block.
Germline mutation testing is performed on FFPE block and peripheral white cells from the EDTA blood sample, and is also supplemented by MLPA.
Sequencing : The AmpliSeq for Illumina BRCA Panel is a targeted resequencing assay for somatic and germline variants across BRCA1 and BRCA2 genes, targeting all the coding exonic regions and the flanking intronic sequences of the BRCA1 and BRCA2 genes.
MLPA : SALSA MLPA Probemix kits (MRC, Holland):
P002 BRCA1, and
P090 BRCA2;
confirmed by
P087 BRCA1 Confirmation or
P077 BRCA2 Confirmation whichever is applicable;
Coffalyzer.Net software v.220513.1739 (database v.220401.0000) for quality control, visualization and analysis of data.
**MLPA using P239 Kit BRCA1 region was retired in Feb-2025, to reduce labour/cost and due to the low yield.
All findings are to be confirmed by orthogonal methods, including Sanger sequencing, RT-PCR and FISH. Germline mutations are ideally confirmed in two tissue types.
Consent is necessary for ordering this test (either somatic test alone, or somatic and germline tests).
For ovarian cancers (Engish) (Chinese)
For breast cancers (Engish) (Chinese)
Pamphlets for BRCA testing in high staged, high grade ovarian cancer (English)(Chinese)
Pamphlets for BRCA testing in breast cancer (English)(Chinese)
The Oncomine Precision Assay, Ion Torrent Genexus System, is an automatic targeted assay for genetic analysis of 50 genes with known relevance to solid tumors. Variants can be confirmed by orthogonal methods (PCR-Sanger sequencing, RT-PCR + Sanger, FISH, etc.). SNV/indel, CNV and fusion variants are included.
Actionable variants in a 12-gene lung cancer somatic mutation panel are reported:
ALK, BRAF, EGFR, ERBB2, KRAS, MET, NTRK1, NTRK2, NTRK3, PIK3CA, RET and ROS1 genes
Hot spot mutations (45):
AKT1 AKT2 AKT3 ALK AR ARAF BRAF CDK4 CDKN2A CHEK2 CTNNB1 EGFR ERBB2 ERBB3 ERBB4 ESR1 FGFR1 FGFR2 FGFR3 FGFR4 FLT3 GNA11 GNAQ GNAS HRAS IDH1 IDH2 KIT KRAS MAP2K1 MAP2K2 MET MTOR NRAS NTRK1 NTRK2 NTRK3 PDGFRA PIK3CA PTEN RAF1 RET ROS1 SMO TP53
Copy Number Variants (14):
ALK AR CD274 CDKN2A EGFR ERBB2 ERBB3 FGFR1 FGFR2 FGFR3 KRAS MET PIK3CA PTEN
Fusion Drivers (18):
ALK AR BRAF EGFR ESR1 FGFR1 FGFR2 FGFR3 MET NRG1 NTRK1 NTRK2 NTRK3 NUTM1 RET ROS1 RSPO2 RSPO3
